February 2023 Progress Update
Screening at scale
At NewLimit, we’re developing a new class of epigenetic reprogramming medicines to treat age-related diseases. At the core of our approach is a Discovery Engine that allows us to search for combinations of reprogramming factors that restore healthy, youthful cell function in aged and diseased cells.
In February, we demonstrated both experimental and in silico portions of our Engine for the first time.
Why share progress updates?
NewLimit is pursuing a more open communications strategy than is common in biotech. We hope that scientists, engineers, and builders of all stripes can benefit from watching how we work.
While moving forward on our scientific goals, we’ve also continued to grow the NewLimit Team. This month we’re excited to welcome two new team members:
Burak Dura as a Principal Scientist leading our single cell genomics efforts
Erin Abernathy as an Executive Assistant on our Operations team
A few highlights on our progress in the world atoms and the world of bits.
Atoms
High-throughput testing of partial reprogramming interventions
For the first time this month, we tested a diverse set of partial reprogramming interventions using our Discovery Engine. To our knowledge, this is the largest partial reprogramming screen to date.
We were excited to confirm that our Engine reproduced the effects of known reprogramming combinations and revealed novel combinations of reprogramming factors with unique phenotypes.
Our first experiments were conducted in a tractable model system. In the coming months, we'll be translating these results to primary T cells for therapeutic discovery.

Expanding the palette of partial reprogramming factors
In our initial experiments, we’ve used a library of reprogramming factors constructed in 2022. This library was always a starting line, rather than a destination.
This month, our Epigenetic Editing team led the charge on increasing the number of colors in our metaphorical palette.
Leveraging an in-house library of human transcription factors, we increased the number of reprogramming factors in our new libraries by >50%.
Learning how not to manufacture reprogramming libraries
One of the key steps in our Discovery Engine is building genetic libraries of reprogramming factor perturbations. By delivering these libraries to cells of interest, we can test many reprogramming combinations in the same dish.
Manufacturing these libraries is non-trivial however! In past months, we've made substantial improvements to this process -- overall, we are now >100X more efficient than our initial manufacturing runs in 2022.
This month we tested another promising idea from the literature to improve manufacturing efficiency and convinced ourselves that the results did not reproduce in our own hands.
While this is disappointing, it's also valuable. NewLimit has a focused, truth-seeking culture. Testing ideas quickly with "killer experiments" and abiding by the results is one of the best mechanisms to ensure our resources are allocated where they're most valuable.
Bits
In silico screening of reprogramming combinations
While our Discovery Engine ramps up, our Predictive Modeling team has made steady progress on building in silico models of reprogramming effects using foundational datasets in the public domain.
This month, we built our first models that can predict the effects of unseen reprogramming combinations on T cell function in a complete reprogramming setting. These data are an imperfect but useful proxy for the partial reprogramming interventions we're developing.
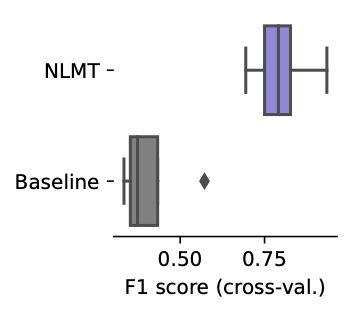
We were excited to find that even our initial approaches are able to outperform baseline models that assume additive effects, suggesting that we can capture non-linear TF interactions and model their effects in silico.

Prospecting the epigenetic landscape
In late 2022, we generated epigenetic maps of aging in primary human T cells. These maps allow us to both (1) build in silico assays for cellular age and (2) to generate hypotheses for reprogramming interventions.
This month, we built robust assays for cellular age prediction based on representation learning models for cell identity prediction. We found that even in the most conservative settings, the single cell genomics measurements collected by our Discovery Platform were sufficient to discriminate cell ages.
Join the team
NewLimit is pursuing an ambitious mission at a fast pace. We’re continuing to expand our team across all of our functional domains. If you’re excited by our mission, please consider applying to NewLimit and reaching out.

