September-October 2023 Progress Update
Expanding the palette & compounding returns
Over the summer, we improved our screening technology and executed our first large scale partial reprogramming screens. We’ve been heads-down for the first months of autumn applying these tools to our first experiments aimed squarely at therapeutic discovery.
These preparations have yielded a series of improvements to our Discovery Engine, which we are continually looking to scale. Each of these iterations compounds with those before it, so that even small wins, month over month, have improved how quickly we can search the space of new sets of reprogramming factors. We look forward to sharing highlights from our larger screens later this year.
People
We’ve had the privilege of welcoming four new team members to NewLimit over the past two months:
Sophie Liu joined as a Research Associate on the Write team.
Neal Ravindra joined as a Machine Learning Scientist on the Predict team.
Jordan Spice joined as a Research Associate on the Write team.
Sarah You joined as an Operations Assistant on the Operations team.
Atoms
Expanding the palette of partial reprogramming factors
Near the end of the summer, we began constructing a partial reprogramming factor library that was 6X larger than any we had built to date. This month, we completed that construction process and successfully completed our first discovery screen using these new factors.
To the best of our knowledge, this is the largest partial reprogramming library that has been screened to date, and this experiment alone more than 3Xs the number of partial reprogramming factors that have been tested for their impact on cell age.

Raising signal above the noise
Our Discovery Engine delivers a pool of reprogramming factors to a pool of cells. Based on the stochasticity of life, each cell receives a different combination of factors from the pool. We then profile cells with single cell genomics so that each cell provides it’s own independent datapoint. This approach allows us to screen many possible combinations in a single culture dish.
One challenge with this approach is that we need to determine which factors are in which cells post-hoc at high fidelity. It turns out this is a rather hard problem! We’ve built a system of DNA barcodes and a companion single cell genomics assay to perform this demultiplexing step during our screening process.
This month, we were able to make a dramatic improvement in the signal-to-noise ratio of that demultiplexing assay. In practice, this means that we can perform screens with more complex combinations of reprogramming factors in each cell while maintaining high fidelity assignment of reprogramming factor combinations to cells.
Enabling ensemble screens for partial reprogramming factors
Most of our screening efforts to date have employed single cell profiling as the endpoint. For each set of reprogramming factors in each cell, we’ve measured a complex phenotype like the expression of every gene, or the accessibility of every region in the genome. These assays are powerful and we can scale them to thousands of factor sets per experiment.
In order to screen tens of thousands of factor sets at the same time though, we’re building an assay that makes different trade-offs. Rather than collecting a complex profile as the endpoint, we simply measure the abundance of each combination of factors in a pool of cells. If we pair this simple measurement with some way to select for cells that look or act more youthful after reprogramming, we can interrogate the effect of many combinations on this selectable feature at an even larger scale than our single cell screens.
This general approach is known as ensemble (“bulk”) screening, and it has proven incredibly valuable in the broader functional genomics community. However, a key challenge we face is that existing methods are unable to detect if reprogramming factors are acting in combination!
Given the importance of combinatorial logic for reprogramming, we’ve been developing a custom next generation sequencing assay to measure the abundance of reprogramming factor combinations in an ensemble format. Our first v0 approach slightly improved on our ability to detect combinations relative to existing methods, but sacrificed our ability to detect single factors in each cell accurately.
This month, we deployed our v1 approach that matches the single factor detection performance of a baseline and achieves much higher performance for detecting factor combinations. We’ll be putting this new assay into production in the coming months.

Increasing Discovery Engine RPM
Our Discovery Engine implements a design-build-test-learn cycle in which we test reprogramming factor combinations for their ability to reverse cell age, then learn from the results to suggest new factors to test.
One of our high level hypotheses is that repeating this cycle more times will increase our chances of success. This month, we focused on shortening the timeline required to execute one of our reprogramming discovery screens in order to fit more engine cycles into each unit of time. Our team managed to reduce the time per cycle by 40%, commensurately increasing our Engine RPM by 1.6X.
Bits
In silico reprogramming at higher fidelity
We’ve been developing in silico reprograming models to predict the effect of a reprogramming factor set on cell age since the beginning of 2023. Over the past two months, we’ve made meaningful improvements to these models that increase the correlation between observed and predicted reprogramming effects. At this stage, we believe our models can predict reprogramming effects as well or better than the prior state-of-the-art (SOTA).

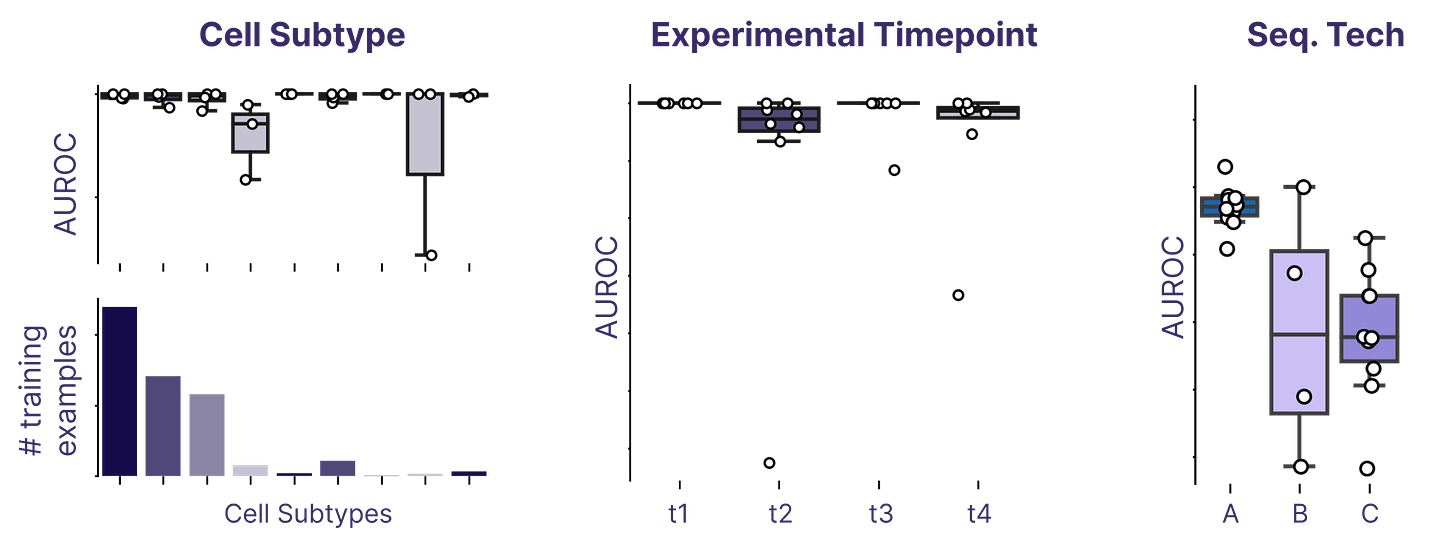
Inferring cell age across diverse domains
Earlier this year, we developed machine learning models that learn to infer cell age — the age of the organism the cell came from — from cell profiles. We’ve previously observed strong performance across a range of conditions, but in the past two months, we spent more time than ever putting these models through their paces and evaluating robustness.
Biological data frequently suffers from domain shifts (aka “batch effects”) where the target data for predictions is a bit different than any of the training data. This can make it challenging to develop models that are robust against technical changes and consistently provide high performance. We put our models through a gamut of challenging domain shift tasks, including predicting on cells from different subtypes, predicting age across experimental conditions, and predicting on entirely distinct sequencing technologies.
We were happy to find that our models performed well, in spite of all the challenges we threw at them. Experiments like these are essential to build confidence in our in silico phenotyping tools, and we’ll continue to act as our own red team against our models in the days to come.